The Simsen Process
The industry leading Simsen process is a pinnacle of versatility
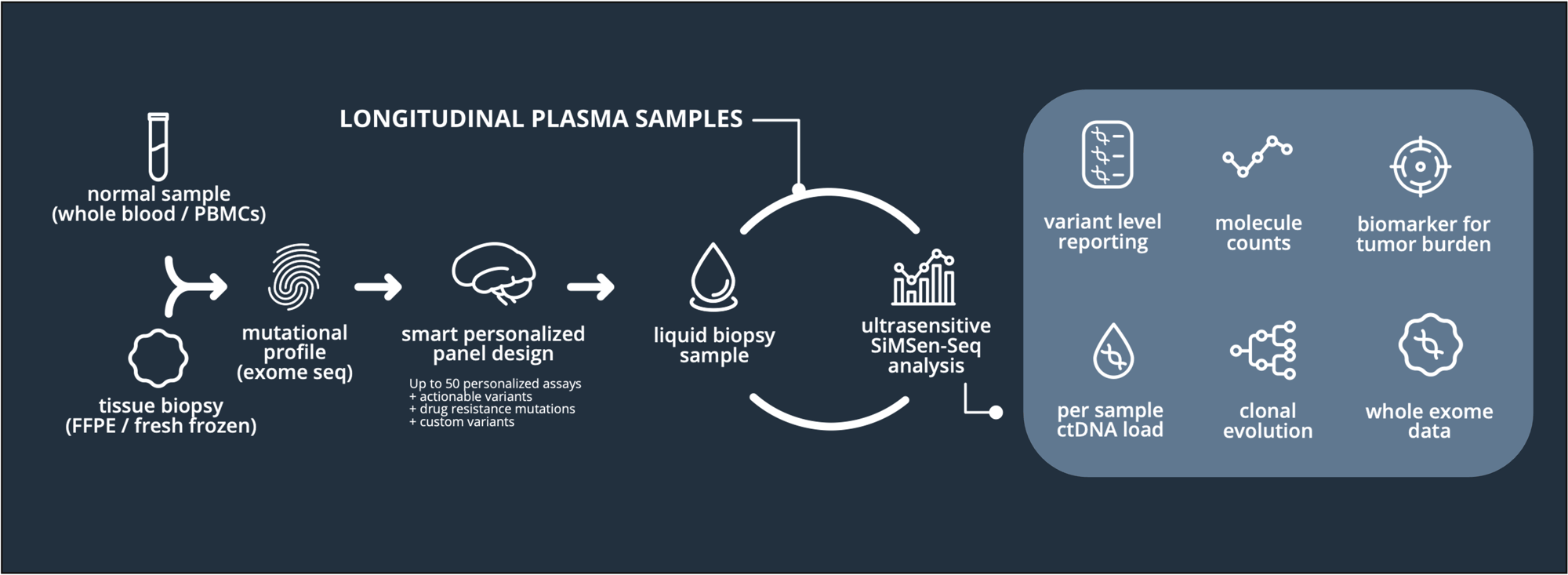
Overview
Simsen Diagnostics will extract genomic DNA from tumor material, which the customer provides either as formalin-fixed, paraffin-embedded tissue (FFPE), or fresh-frozen tumor tissue. The extracted DNA will then be subjected to exome sequencing by Simsen Diagnostics or by a 3rd party sequencing provider with whom Simsen Diagnostics has a quality agreement.
Formalin-fixed, paraffin-embedded tissue material presents challenges not only for the DNA extraction method itself but also for the determination of DNA quality and quantity. Generally, DNA yield from FFPE samples varies greatly, depending on the tissue type and fixation and embedding conditions. Fresh frozen tissue samples are preferable over FFPE for obtaining high-quality DNA.
Ideally, tissue samples will be complemented by a normal control sample (e.g., a matched sample of whole blood) to remove false positive somatic mutations and increase the number of high-quality tumor-specific mutations. Tumor-only variant calling is more bioinformatically challenging and will likely result in reduced sensitivity of the final ctDNA analysis due to a much smaller number of identified targets. Thus, matched tumor-normal analysis is highly recommended.
In the case of FFPE tissue, DNA will be extracted using the QIAamp DNA FFPE Advanced kit from Qiagen.
In the case of fresh frozen tissue, DNA will be extracted using the QIAamp DNA Mini / Blood Mini Kit from Qiagen.
The Blood Mini Kit will also be used for the extraction of matched normal blood samples.
Shipping and storage of tissue material
FFPE tissue in tubes or on slides can be sent at room temperature. Storage prior to extraction will be at 4℃.
Fresh frozen tissue and whole blood samples should be sent on dry ice and will be stored at -80℃ prior to extraction.
Sample requirements
Simsen Tumor Profiling
Tumor tissue
| Tissue source | Recommended input | Alternative input1 | Shipping recommendations |
| Fresh frozen | 10-30 mg | > 1 mg | Dry ice |
| FFPE2 | 10 fresh sections / curls with 10 - 20 µm thickness or equivalent (e.g. 20 sections of 5 µm thickness) | > 2 curls of > 5 µm thickness or 10 - 20 unstained glass slides with fixed FFPE sections | Ambient |
| Extracted DNA3 | > 250 ng at > 10 ng/µl | < 50 ng at > 1 ng/µl | Dry ice |
[1] Alternative values indicate the minimum amount required to likely achieve a satisfactory result.
[2] Tumor content must be >30%, as assessed by the customer. We currently do not accept whole tumor blocks!
[3] DNA should be delivered in molecular biology grade water, 10 mM Tris-HCl pH 8 or EB, free of EDTA. Buffers containing high amounts of EDTA may cause downstream inhibition.
Germline control
It is highly recommended to use a matched germline control for each subject (“normal sample”) in conjunction with the tumor tissue for accurate somatic variant calling. Typically, whole blood collected in EDTA tubes is the most convenient source of matched control DNA.
Simsen Personal Collection kits contain two Streck preservative tubes for ctDNA analysis and a single EDTA tube for providing a matched germline control sample. The collection kits can be stored a shipped at ambient temperature using the contained return label. The EDTA must only be provided once per patient, typically at the first blood draw for ctDNA analysis together with the Streck tubes.
| Sample type | Recommended input | Alternative input | Shipping recommendation |
| Whole blood (EDTA) | 1 ml | > 100 µl | Dry ice |
| Cell pellet from whole blood | > 1 million cells | > 100 000 cells | Dry ice |
| Dried blood spots on FTA filter cards | 5 spots > 100 µl | 1 spot > 100 µl | Ambient |
| Extracted DNA | > 250 ng at > 10 ng/µl | > 50 ng at >1 ng/µl | Dry ice |
| Simsen Personal Collection Kit | Whole blood in 6 ml EDTA tube | - | Ambient |
Other sources of control DNA, e.g. from tissues other than blood or other formats such as fresh frozen or FFPE may also be used.
Customer provided mutational profiles
It is possible to use already existing genomic data, instead of providing tissue material, and provide a mutational profile to Simsen, please consider the following to achieve optimal results:
- Provide at least 5, preferably 20 – 30 mutations. The sensitivity of the analysis is directly proportional to the number of variants analyzed. Fewer mutations are possible. More mutations also allow us more flexibility when designing the personalized panel.
- Mutations must be accessible on cell-free DNA by PCR (SNV, short indels, specific breakpoints)
- Use best practices for variant calling and sequencing (e.g., using a matched normal control)
- Use high quality tumor tissue for sequencing
- Use tumor tissue that is most representative of the current disease state
- Any method capable of accurate somatic mutation detection (e.g. TSO 500, WES, WGS, etc.) is suitable for detecting variants that can be followed in liquid biopsy samples using Simsen Personal
Acceptable data formats
Target variants can be provided in a simple spread sheet or VCF file. If raw sequencing data in fastq format is provided, we can process it using our proprietary variant calling pipeline.
We cannot guarantee that mutations provided by customers are indeed somatic tumor mutations and thus can be detected in the circulation.
For inquiries about customer provided mutational profiles and data, please contact support@simsendiagnostics.com.
Optional Add-on Variants
Whether you choose to provide us with tumor tissue for mutational profiling or provide your own tumor mutational profile for personalized variant tracking, we can add custom variants to personalized panels, including:
- Pre-defined driver and resistance mutations
- Other variants (e.g., neoantigens)
- Custom generic ultrasensitive targeted amplicon panels (with or without personalized variants)
Other sample types
Archival materials, e.g., from biobanks, can be used if the amount and quality of the material is in line with criteria outlined above. We have extensive experience working with limited and low-quality sample. For inquiries about sample types not listed above, please contact support@simsendiagnostics.com.
ctDNA analysis / liquid biopsy
Sample requirements
| Tissue source | Recommended input | Alternative input | Shipping recommendations |
| Plasma | 10 ml | > 1 ml | Dry ice |
| cfDNA | > 25 ng | > 5 ng | Dry ice |
| Whole blood in preservative tubes | 2 tubes | 1 tube | Ambient |
If the sample handling times for EDTA cannot be met, preservative tubes may be used instead. Our own collection kits use Streck BCT tubes, but other commonly used tubes such as PaxGene, Norgen or Roche cfDNA collection tubes are also acceptable. Use only preservative tube dedicated for use with circulating cell-free DNA. Preserved blood samples can be shipped at ambient temperature and are stable for several days or even weeks. If using preservative tubes, please follow the manufacturers instruction for collection, storage and shipment.
Simsen Personal collection kits
Simsen Personal Collection kits contain two Streck preservative tubes for ctDNA analysis and a single EDTA tube for providing a matched germline control sample. The collection kits can be stored and shipped at ambient temperature using the contained return label.
Other liquid biopsy samples
It is possible to accommodate sample types other than those described above.
- Most other blood collection tubes, such as citrate, will likely work but have not been tested by us.
- Do not use blood collection tubes containing heparin!
- Serum samples are acceptable, but data must be interpreted with care, as serum samples contain increased levels of wildtype DNA, which artificially deflates variant allele frequencies.
- Alternative sample types such as cerebrospinal fluid, urine, sputum or saliva can be used but must be discussed with Simsen Diagnostics to ensure that sample quality is high enough for successful analysis.
Reporting
-
Requirements and recommendations
Reporting overview
 We provide comprehensive reports with all our analysis, including sharing of all raw data via secure file transfer.
We provide comprehensive reports with all our analysis, including sharing of all raw data via secure file transfer.
Our ctDNA reports contain the following metrics for each sample:
- Mutant molecules per mL plasma (MM/mL)
- Variant allele frequency (VAF)
- Amount of cell-free DNA used
Both MM/mL and VAF are reported as a single value for each sample, the ‘ctDNA load’ and for all personalized variants individually, allowing the tracking of clonal evolution. Each report also contains a ctDNA timeline of all available ctDNA measurements from the same subject.
To view an example report, please click below ↓
If Simsen Diagnostics performed exome sequencing and mutational profile identification, that data will also be reported.
Which metrics should be used?
MM/mL plasma and VAF are frequently used to report ctDNA results. Changes in ctDNA over time will in many cases be highly correlated between MM/mL and VAF. However, in discordant cases, that is, where ctDNA levels change from timepoint A to B in one metric but not the other, MM/mL has been shown to have a stronger correlation with clinical outcomes. Furthermore, transient increases in (wildtype) cell-free DNA can be due to a number of physiological and preanalytical factors, which may artificially decrease the allele fraction, while MM/mL remain unchanged. Nevertheless, we favor reporting both results for maximum transparency, but based on currently available evidence and best practices recommend using MM/mL for correlation with clinical outcomes.
